Pancreatic islet chromatin accessibility and conformation defines distal enhancer networks of type 2 diabetes risk
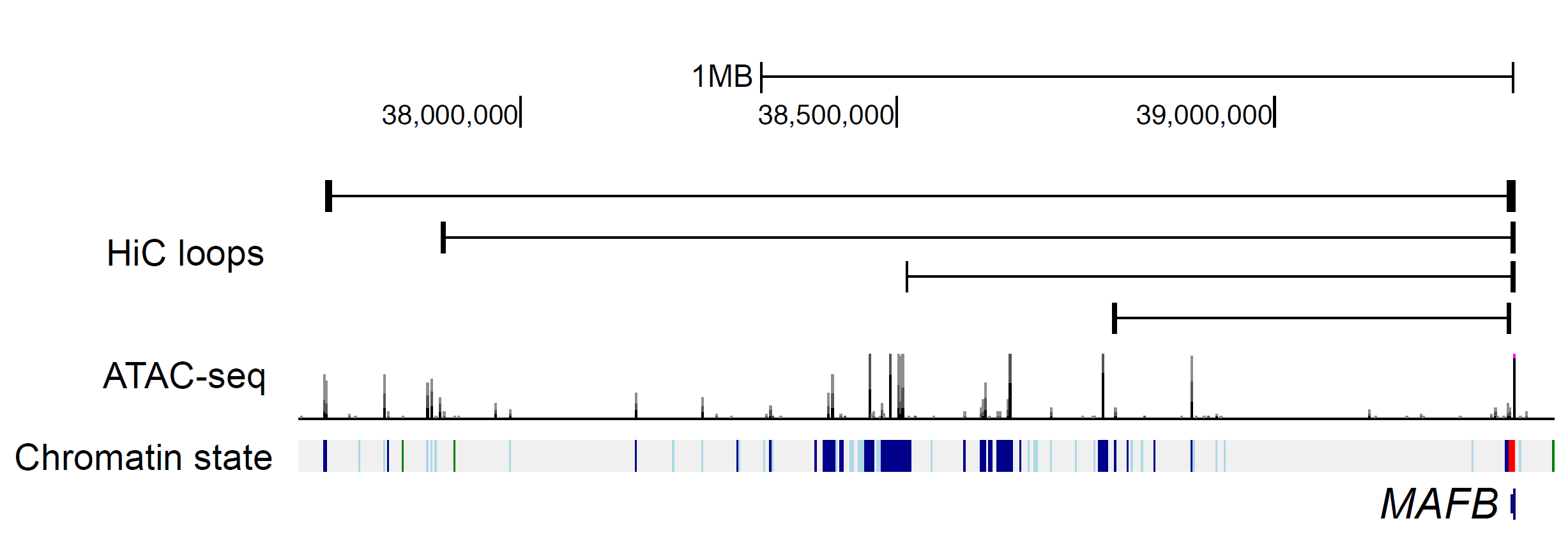 Figure 2B: Islet enhancer loops between MAFB and distal enhancers
Figure 2B: Islet enhancer loops between MAFB and distal enhancers
Citation: William W Greenwald*, Joshua Chiou*, Jian Yan*, Yunjiang Qiu*, Ning Dai, Allen Wang, Naoki Nariai, Anthony Aylward, Jee Yun Han, Nikita Kadakia, Laura Barrufet, Mei-Lin Okino, Frauke Drees, Nicholas Vinckier, Liliana Minichiello, David Gorkin, Joseph Avruch, Kelly Frazer, Maike Sander, Bing Ren, Kyle J Gaulton. (2018) Pancreatic islet chromatin accessibility and conformation defines distal enhancer networks of type 2 diabetes risk. bioRxiv.
doi: https://doi.org/10.1101/299388
* Authors contributed equally to this work
Islet epigenome data
Data description:
- ATAC-seq peaks merged from 4 samples generated in this study
- Hi-C loops merged from 3 samples generated in this study
- Chromatin states (8 states) with ChIP-seq data from Parker et al., 2013 and Pasquali et al., 2014
Data availability:
[ATAC-seq peaks] | md5sum: 1f0a20c9ecf15992fc2090a00b045a76
[Hi-C loops] | md5sum: 09e71249b380641dc87db06598be0e11
[Chromatin states] | md5sum: b34b262613c8151286518e91a6ab488c
Last updated:
Josh Chiou | 2018-04-19T05:27:09+00:00